🌽 Crop Classification and Clustering¶
In [ ]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.gridspec as gridspec
import math
## Models
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.model_selection import train_test_split
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
from sklearn.metrics import confusion_matrix, classification_report, accuracy_score
from sklearn.metrics import precision_score, recall_score, f1_score
from sklearn.impute import SimpleImputer
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
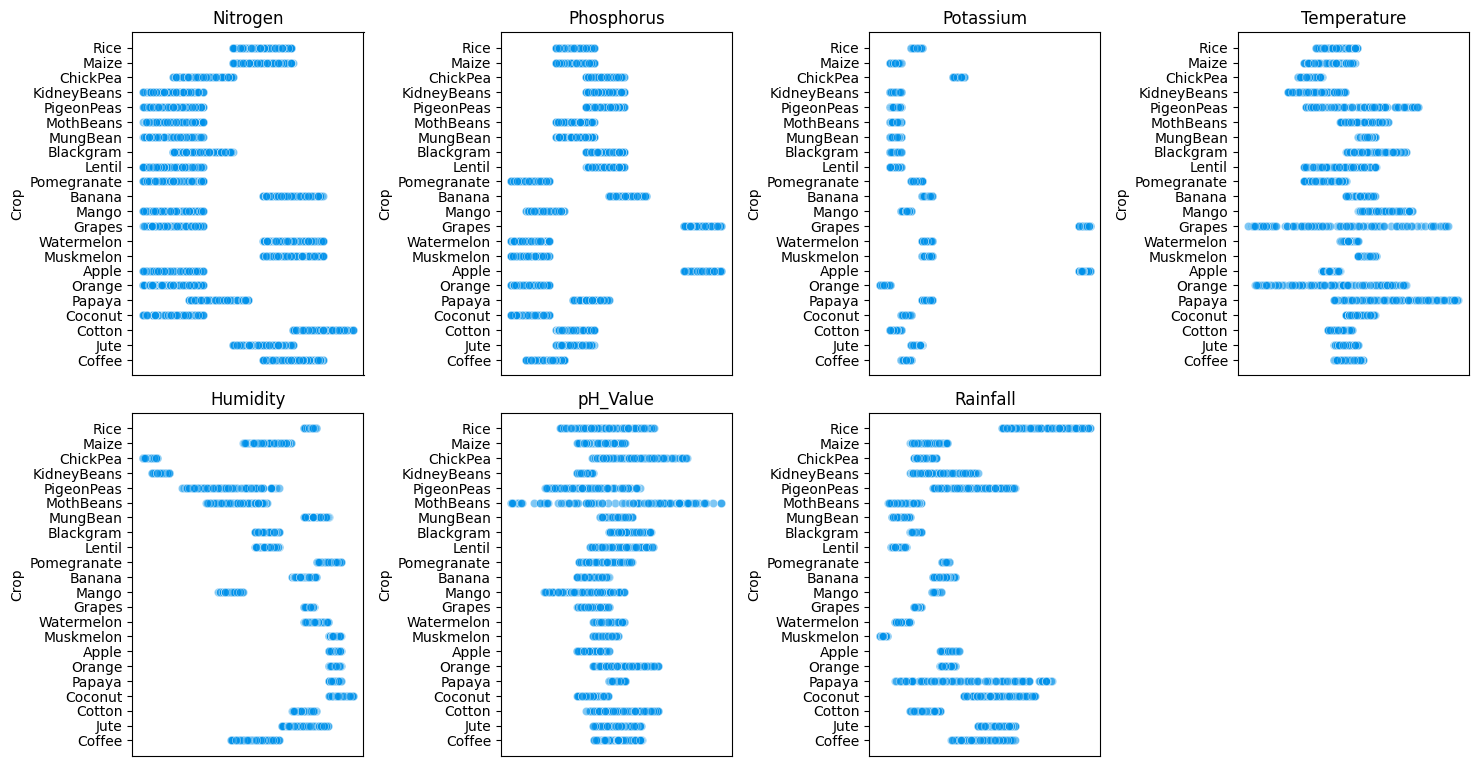
EDA¶
In [ ]:
df = pd.read_csv('Crop_Recommendation.csv')
df
Out[ ]:
| Nitrogen | Phosphorus | Potassium | Temperature | Humidity | pH_Value | Rainfall | Crop | |
|---|---|---|---|---|---|---|---|---|
| 0 | 90 | 42 | 43 | 20.879744 | 82.002744 | 6.502985 | 202.935536 | Rice |
| 1 | 85 | 58 | 41 | 21.770462 | 80.319644 | 7.038096 | 226.655537 | Rice |
| 2 | 60 | 55 | 44 | 23.004459 | 82.320763 | 7.840207 | 263.964248 | Rice |
| 3 | 74 | 35 | 40 | 26.491096 | 80.158363 | 6.980401 | 242.864034 | Rice |
| 4 | 78 | 42 | 42 | 20.130175 | 81.604873 | 7.628473 | 262.717340 | Rice |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2195 | 107 | 34 | 32 | 26.774637 | 66.413269 | 6.780064 | 177.774507 | Coffee |
| 2196 | 99 | 15 | 27 | 27.417112 | 56.636362 | 6.086922 | 127.924610 | Coffee |
| 2197 | 118 | 33 | 30 | 24.131797 | 67.225123 | 6.362608 | 173.322839 | Coffee |
| 2198 | 117 | 32 | 34 | 26.272418 | 52.127394 | 6.758793 | 127.175293 | Coffee |
| 2199 | 104 | 18 | 30 | 23.603016 | 60.396475 | 6.779833 | 140.937041 | Coffee |
2200 rows × 8 columns
In [ ]:
features = df.columns[:-1]
features
Out[ ]:
Index(['Nitrogen', 'Phosphorus', 'Potassium', 'Temperature', 'Humidity',
'pH_Value', 'Rainfall'],
dtype='object')
In [ ]:
df.Crop.nunique()
Out[ ]:
22
In [ ]:
df.Crop.value_counts()
Out[ ]:
Crop Rice 100 Maize 100 Jute 100 Cotton 100 Coconut 100 Papaya 100 Orange 100 Apple 100 Muskmelon 100 Watermelon 100 Grapes 100 Mango 100 Banana 100 Pomegranate 100 Lentil 100 Blackgram 100 MungBean 100 MothBeans 100 PigeonPeas 100 KidneyBeans 100 ChickPea 100 Coffee 100 Name: count, dtype: int64
In [ ]:
df.Crop.value_counts(normalize=True)
Out[ ]:
Crop Rice 0.045455 Maize 0.045455 Jute 0.045455 Cotton 0.045455 Coconut 0.045455 Papaya 0.045455 Orange 0.045455 Apple 0.045455 Muskmelon 0.045455 Watermelon 0.045455 Grapes 0.045455 Mango 0.045455 Banana 0.045455 Pomegranate 0.045455 Lentil 0.045455 Blackgram 0.045455 MungBean 0.045455 MothBeans 0.045455 PigeonPeas 0.045455 KidneyBeans 0.045455 ChickPea 0.045455 Coffee 0.045455 Name: proportion, dtype: float64
In [ ]:
df.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 2200 entries, 0 to 2199 Data columns (total 8 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Nitrogen 2200 non-null int64 1 Phosphorus 2200 non-null int64 2 Potassium 2200 non-null int64 3 Temperature 2200 non-null float64 4 Humidity 2200 non-null float64 5 pH_Value 2200 non-null float64 6 Rainfall 2200 non-null float64 7 Crop 2200 non-null object dtypes: float64(4), int64(3), object(1) memory usage: 137.6+ KB
In [ ]:
df.isna().sum()
Out[ ]:
Nitrogen 0 Phosphorus 0 Potassium 0 Temperature 0 Humidity 0 pH_Value 0 Rainfall 0 Crop 0 dtype: int64
In [ ]:
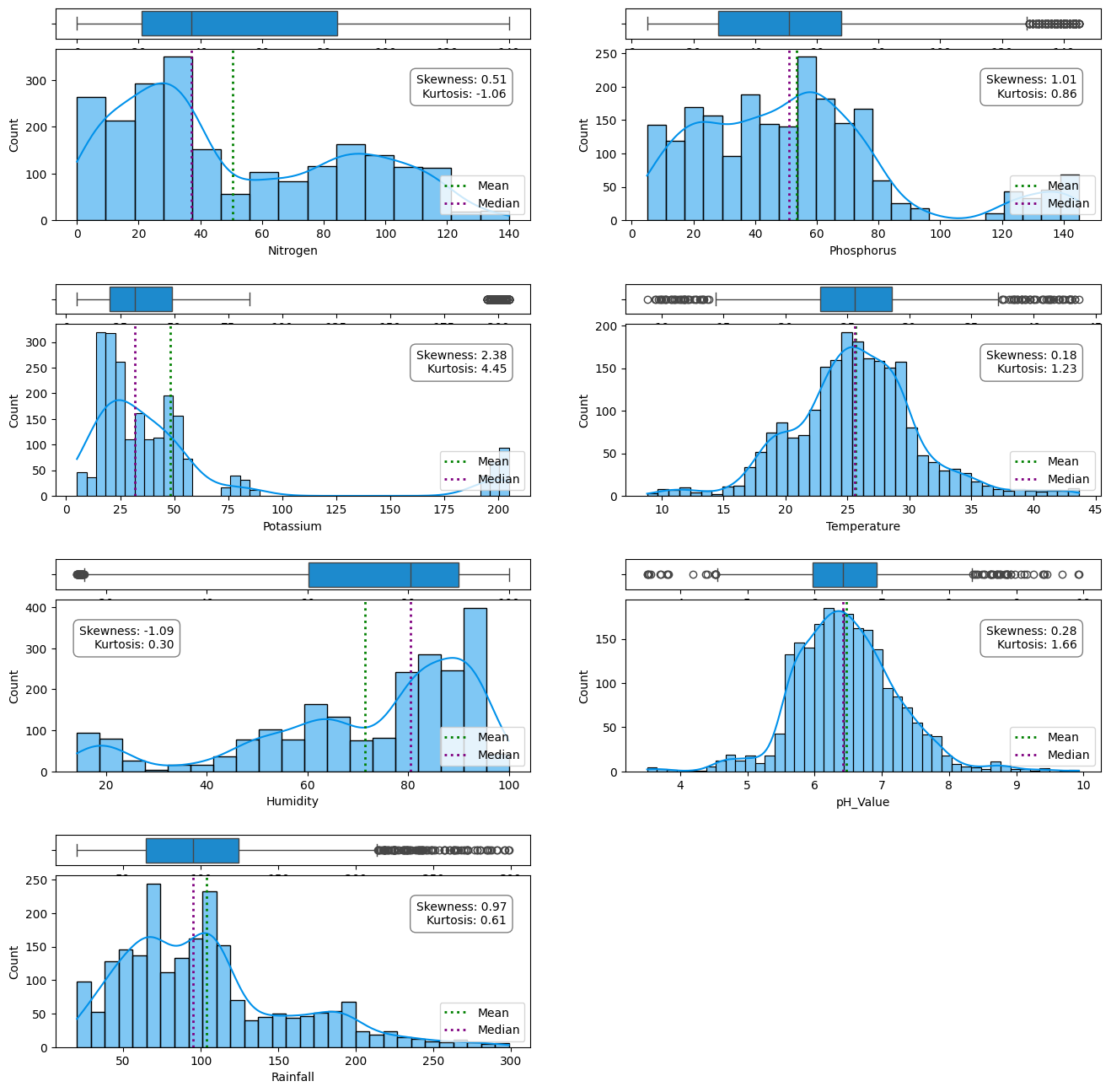
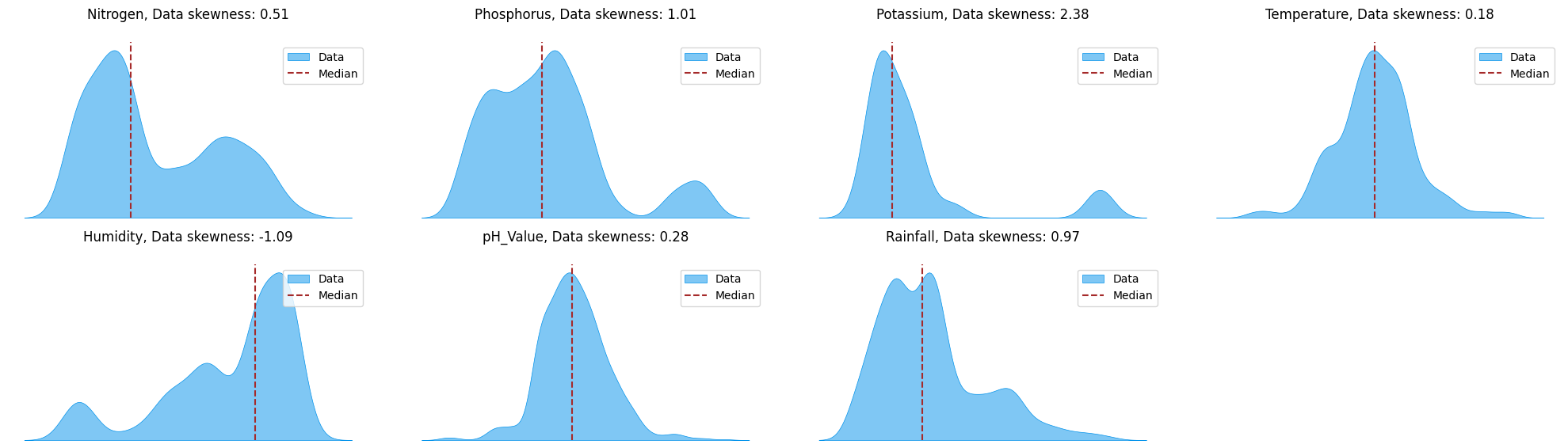
df.describe().T
Out[ ]:
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| Nitrogen | 2200.0 | 50.551818 | 36.917334 | 0.000000 | 21.000000 | 37.000000 | 84.250000 | 140.000000 |
| Phosphorus | 2200.0 | 53.362727 | 32.985883 | 5.000000 | 28.000000 | 51.000000 | 68.000000 | 145.000000 |
| Potassium | 2200.0 | 48.149091 | 50.647931 | 5.000000 | 20.000000 | 32.000000 | 49.000000 | 205.000000 |
| Temperature | 2200.0 | 25.616244 | 5.063749 | 8.825675 | 22.769375 | 25.598693 | 28.561654 | 43.675493 |
| Humidity | 2200.0 | 71.481779 | 22.263812 | 14.258040 | 60.261953 | 80.473146 | 89.948771 | 99.981876 |
| pH_Value | 2200.0 | 6.469480 | 0.773938 | 3.504752 | 5.971693 | 6.425045 | 6.923643 | 9.935091 |
| Rainfall | 2200.0 | 103.463655 | 54.958389 | 20.211267 | 64.551686 | 94.867624 | 124.267508 | 298.560117 |
Probably a good idea to standardize these values
In [ ]:
# Correlation Matrix
corr_matrix = df.corr(numeric_only=True)
plt.figure(figsize=(15, 10))
sns.heatmap(corr_matrix,
annot=True,
linewidths=0.5,
fmt= ".2f",
cmap="YlGnBu");
In [ ]:
metric_col = df.select_dtypes(include=['number']).columns
Modeling¶
In [ ]:
X = df.drop('Crop', axis=1)
y = df.Crop.values
In [ ]:
scaler = StandardScaler()
X = scaler.fit_transform(X)
In [ ]:
# Train and Test Split
np.random.seed(42)
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.2)
In [ ]:
X_train.shape, X_test.shape, y_train.shape, y_test.shape
Out[ ]:
((1760, 7), (440, 7), (1760,), (440,))
In [ ]:
# Put models in a dictionary
models = {"KNN": KNeighborsClassifier(),
"Logistic Regression": LogisticRegression(),
"Random Forest": RandomForestClassifier(),
"GradientBoost": GradientBoostingClassifier(),
"GaussianNB": GaussianNB(),
}
In [ ]:
# Create function to fit and score models
def fit_and_score(models, X_train, X_test, y_train, y_test):
"""
Fits and evaluates given machine learning models.
models : a dict of different Scikit-Learn machine learning models
X_train : training data
X_test : testing data
y_train : labels assosciated with training data
y_test : labels assosciated with test data
"""
# Random seed for reproducible results
np.random.seed(42)
# Make a list to keep model scores
model_scores = {}
# Loop through models
for name, model in models.items():
# Fit the model to the data
model.fit(X_train, y_train)
# Evaluate the model and append its score to model_scores
model_scores[name] = model.score(X_test, y_test)
return model_scores
In [ ]:
model_scores = fit_and_score(models=models,
X_train=X_train,
X_test=X_test,
y_train=y_train,
y_test=y_test)
model_scores
C:\Users\tuckerd9\AppData\Roaming\Python\Python39\site-packages\sklearn\linear_model\_logistic.py:444: ConvergenceWarning: lbfgs failed to converge (status=1):
STOP: TOTAL NO. of ITERATIONS REACHED LIMIT.
Increase the number of iterations (max_iter) or scale the data as shown in:
https://scikit-learn.org/stable/modules/preprocessing.html
Please also refer to the documentation for alternative solver options:
https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression
n_iter_i = _check_optimize_result(
Out[ ]:
{'KNN': 0.9568181818181818,
'Logistic Regression': 0.9636363636363636,
'Random Forest': 0.9931818181818182,
'GradientBoost': 0.9818181818181818,
'GaussianNB': 0.9954545454545455}
I could go with GaussianNB, but just choosing RandomForest model
In [ ]:
model_compare = pd.DataFrame(model_scores, index=['accuracy'])
model_compare.T.plot.bar();
In [ ]:
rf = RandomForestClassifier()
rf.fit(X_train, y_train)
y_preds_rf = rf.predict(X_test)
In [ ]:
# Create a new dataframe with two columns: 'test' and 'predicted'
df_with_preds = pd.DataFrame({
'test': y_test,
'predicted': y_preds_rf
})
df_with_preds
Out[ ]:
| test | predicted | |
|---|---|---|
| 0 | Muskmelon | Muskmelon |
| 1 | Watermelon | Watermelon |
| 2 | Papaya | Papaya |
| 3 | Papaya | Papaya |
| 4 | Apple | Apple |
| ... | ... | ... |
| 435 | Rice | Rice |
| 436 | Rice | Rice |
| 437 | Cotton | Cotton |
| 438 | Cotton | Cotton |
| 439 | PigeonPeas | PigeonPeas |
440 rows × 2 columns
In [ ]:
# Create a new dataframe with the predicted values as a column
df_with_preds = pd.DataFrame(X_test, columns=X_test.columns)
df_with_preds['actual'] = y_test
df_with_preds['predicted'] = y_preds_rf
df_with_preds
Out[ ]:
| Nitrogen | Phosphorus | Potassium | Temperature | Humidity | pH_Value | Rainfall | actual | predicted | |
|---|---|---|---|---|---|---|---|---|---|
| 1451 | 101 | 17 | 47 | 29.494014 | 94.729813 | 6.185053 | 26.308209 | Muskmelon | Muskmelon |
| 1334 | 98 | 8 | 51 | 26.179346 | 86.522581 | 6.259336 | 49.430510 | Watermelon | Watermelon |
| 1761 | 59 | 62 | 49 | 43.360515 | 93.351916 | 6.941497 | 114.778071 | Papaya | Papaya |
| 1735 | 44 | 60 | 55 | 34.280461 | 90.555616 | 6.825371 | 98.540477 | Papaya | Papaya |
| 1576 | 30 | 137 | 200 | 22.914300 | 90.704756 | 5.603413 | 118.604465 | Apple | Apple |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 59 | 99 | 55 | 35 | 21.723831 | 80.238990 | 6.501698 | 277.962619 | Rice | Rice |
| 71 | 67 | 45 | 38 | 22.727910 | 82.170688 | 7.300411 | 260.887506 | Rice | Rice |
| 1908 | 121 | 47 | 16 | 23.605640 | 79.295731 | 7.723240 | 72.498009 | Cotton | Cotton |
| 1958 | 116 | 52 | 19 | 22.942767 | 75.371706 | 6.114526 | 67.080226 | Cotton | Cotton |
| 482 | 5 | 68 | 20 | 19.043805 | 33.106951 | 6.121667 | 155.370562 | PigeonPeas | PigeonPeas |
440 rows × 9 columns
In [ ]:
def plot_confusion_matrix(y_test, predictions):
# Plot the confusion matrix
cf_matrix = confusion_matrix(y_test, predictions)
fig = plt.subplots(figsize=(10, 8))
sns.set(font_scale=1.4)
sns.heatmap(cf_matrix, annot=True, fmt='d')
plt.xlabel('Predicted Label', fontsize=12)
plt.xticks(fontsize=12)
plt.ylabel('True Label', fontsize=12)
plt.yticks(fontsize=12)
plt.show()
# Reset font scale to default
sns.set(font_scale=1)
In [ ]:
# Confusion matrix
print(confusion_matrix(y_test, y_preds_rf))
In [ ]:
plot_confusion_matrix(y_test, y_preds_rf)
In [ ]:
# Show classification report
print(classification_report(y_test, y_preds_rf))
precision recall f1-score support
Apple 1.00 1.00 1.00 23
Banana 1.00 1.00 1.00 21
Blackgram 1.00 1.00 1.00 20
ChickPea 1.00 1.00 1.00 26
Coconut 1.00 1.00 1.00 27
Coffee 1.00 1.00 1.00 17
Cotton 1.00 1.00 1.00 17
Grapes 1.00 1.00 1.00 14
Jute 0.92 1.00 0.96 23
KidneyBeans 1.00 1.00 1.00 20
Lentil 0.92 1.00 0.96 11
Maize 1.00 1.00 1.00 21
Mango 1.00 1.00 1.00 19
MothBeans 1.00 0.96 0.98 24
MungBean 1.00 1.00 1.00 19
Muskmelon 1.00 1.00 1.00 17
Orange 1.00 1.00 1.00 14
Papaya 1.00 1.00 1.00 23
PigeonPeas 1.00 1.00 1.00 23
Pomegranate 1.00 1.00 1.00 23
Rice 1.00 0.89 0.94 19
Watermelon 1.00 1.00 1.00 19
accuracy 0.99 440
macro avg 0.99 0.99 0.99 440
weighted avg 0.99 0.99 0.99 440
In [ ]:
# Get feature importance scores
importance_scores = rf.feature_importances_
# Get feature names
feature_names = X_train.columns.tolist()
In [ ]:
# Sort feature importance scores in descending order
sorted_idx = np.argsort(importance_scores)[::-1]
# Create plot
plt.figure(figsize=(10, 8))
# Plot feature importance scores
for i in range(len(importance_scores)):
plt.bar(i, importance_scores[sorted_idx[i]], color='#0091ea', align='center')
plt.text(i,
importance_scores[sorted_idx[i]]+0.01,
feature_names[sorted_idx[i]],
horizontalalignment='center')
# Add title and labels
plt.title('Feature Importance')
plt.xlabel('Feature Index')
Out[ ]:
Text(0.5, 0, 'Feature Index')
Hypermeter Tuning with GridSearchCV¶
In [ ]:
param_grid = {
'n_estimators': [10, 50, 100, 200],
'max_depth': [None, 10, 20, 30]
}
In [ ]:
grid_search = GridSearchCV(
estimator=rf,
param_grid=param_grid,
scoring='accuracy',
cv=5,
verbose=True,
)
grid_search.fit(X_train, y_train)
print("Best parameters: ", grid_search.best_params_)
print("Accuracy score: ", grid_search.score(X_test, y_test))
Fitting 5 folds for each of 16 candidates, totalling 80 fits
Best parameters: {'max_depth': 20, 'n_estimators': 50}
Accuracy score: 0.9931818181818182
- No improvement of the original model with gridsearch
In [ ]:
# Get the best estimator
best_estimator = grid_search.best_estimator_
# Make predictions on the test set
y_pred = best_estimator.predict(X_test)
# Print the predictions
print("Predictions: ", y_pred)
Clustering¶
In [ ]:
df_cluster = df.drop('Crop', axis=1)
In [ ]:
# Not needed for this example since there are no categorical variables
df_cluster = pd.get_dummies(df_cluster, drop_first=True)
df_cluster.head(5)
Out[ ]:
| Nitrogen | Phosphorus | Potassium | Temperature | Humidity | pH_Value | Rainfall | |
|---|---|---|---|---|---|---|---|
| 0 | 90 | 42 | 43 | 20.879744 | 82.002744 | 6.502985 | 202.935536 |
| 1 | 85 | 58 | 41 | 21.770462 | 80.319644 | 7.038096 | 226.655537 |
| 2 | 60 | 55 | 44 | 23.004459 | 82.320763 | 7.840207 | 263.964248 |
| 3 | 74 | 35 | 40 | 26.491096 | 80.158363 | 6.980401 | 242.864034 |
| 4 | 78 | 42 | 42 | 20.130175 | 81.604873 | 7.628473 | 262.717340 |
In [ ]:
# Finding optimal number of clusters
from sklearn.mixture import GaussianMixture
# Prepare
n_components = np.arange(1,10)
# Create GMM model
models = [GaussianMixture(n_components= n,
random_state = 1502).fit(df_cluster) for n in n_components]
#Plot
plt.plot(n_components,
[m.bic(df_cluster) for m in models],
label = 'BIC')
plt.plot(n_components,
[m.aic(df_cluster) for m in models],
label = 'AIC')
plt.legend()
plt.xlabel('Number of Components')
Out[ ]:
Text(0.5, 0, 'Number of Components')
In [ ]:
# Gaussian Mixture Model
model = GaussianMixture(n_components= 4,
random_state = 1502).fit(df_cluster)
In [ ]:
# Predict the cluster for each customer
cluster = pd.Series(model.predict(df_cluster))
cluster[:2]
Out[ ]:
0 1 1 1 dtype: int64
In [ ]:
# Create Cluster variable
df_cluster['cluster'] = cluster
df_cluster.head(5)
Out[ ]:
| Nitrogen | Phosphorus | Potassium | Temperature | Humidity | pH_Value | Rainfall | cluster | |
|---|---|---|---|---|---|---|---|---|
| 0 | 90 | 42 | 43 | 20.879744 | 82.002744 | 6.502985 | 202.935536 | 1 |
| 1 | 85 | 58 | 41 | 21.770462 | 80.319644 | 7.038096 | 226.655537 | 1 |
| 2 | 60 | 55 | 44 | 23.004459 | 82.320763 | 7.840207 | 263.964248 | 1 |
| 3 | 74 | 35 | 40 | 26.491096 | 80.158363 | 6.980401 | 242.864034 | 1 |
| 4 | 78 | 42 | 42 | 20.130175 | 81.604873 | 7.628473 | 262.717340 | 1 |
Visualizing the clustering¶
In [ ]:
from sklearn.decomposition import PCA
In [ ]:
data_pca = df_cluster.drop(['cluster'], axis=1)
3D Scatter of the data
In [ ]:
#Initiating PCA to reduce dimentions aka features to 3
pca = PCA(n_components=3)
pca.fit(data_pca)
PCA_ds = pd.DataFrame(pca.transform(data_pca), columns=(["col1","col2", "col3"]))
PCA_ds.describe().T
Out[ ]:
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| col1 | 2200.0 | -2.480440e-15 | 58.604247 | -104.182669 | -35.757181 | -8.465669 | 10.306607 | 180.711278 |
| col2 | 2200.0 | 7.441320e-15 | 54.157657 | -84.309526 | -44.703712 | -10.190573 | 44.016507 | 170.783030 |
| col3 | 2200.0 | -4.134067e-15 | 36.728830 | -67.731895 | -30.630587 | -7.256216 | 28.947581 | 81.762206 |
In [ ]:
PCA_ds['Cluster'] = df_cluster['cluster']
PCA_ds
Out[ ]:
| col1 | col2 | col3 | Cluster | |
|---|---|---|---|---|
| 0 | -59.969729 | 84.055388 | 32.450240 | 1 |
| 1 | -64.090628 | 107.779037 | 24.381552 | 1 |
| 2 | -75.156888 | 142.468675 | -0.556024 | 1 |
| 3 | -80.247626 | 117.340628 | 13.940485 | 1 |
| 4 | -85.084925 | 137.343003 | 16.712434 | 1 |
| ... | ... | ... | ... | ... |
| 2195 | -64.055404 | 54.019513 | 44.868290 | 1 |
| 2196 | -52.816986 | 3.172884 | 38.516656 | 1 |
| 2197 | -65.984590 | 48.821451 | 55.387287 | 1 |
| 2198 | -42.988902 | 7.978078 | 55.117203 | 1 |
| 2199 | -55.797011 | 16.737268 | 43.669666 | 1 |
2200 rows × 4 columns
In [ ]:
# Export the plot as an HTML file
import plotly.graph_objects as go
import plotly.io as pio
# Write the scatterplot to an HTML file
pio.write_html(fig, file='scatterplot.html')
2D scatterplot of the data¶
In [ ]:
# Visualize the clusters using PCA
pca2 = PCA(n_components=2)
principal_components = pca2.fit_transform(X)
PCA_ds['PCA1'] = principal_components[:, 0]
PCA_ds['PCA2'] = principal_components[:, 1]
# Plot the PCA result
plt.figure(figsize=(10, 8))
sns.scatterplot(data=PCA_ds, x='PCA1', y='PCA2', hue='Cluster', palette='Set1', s=100)
plt.title('PCA of Crop Clusters')
plt.xlabel('PCA Component 1')
plt.ylabel('PCA Component 2')
plt.legend(title='Cluster')
plt.show()
Once I have narrowed down the RandomForest model that I want to utilize: Streamline the process
Combined preprocessing and gridsearchcv in one process
In [ ]:
import pandas as pd
import numpy as np
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV, train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_classification
# Does not apply to this dataset
# categorical_features = df.select_dtypes(exclude=['number']).columns
# categorical_transformer = Pipeline(steps=[
# ("imputer", SimpleImputer(strategy="constant", fill_value="missing")),
# ("onehot", OneHotEncoder(handle_unknown="ignore"))
# ])
# Create a pipeline for numeric features
numeric_features = df.select_dtypes(include=['number']).columns
numeric_transformer = Pipeline(steps=[
("imputer", SimpleImputer(strategy="mean")),
('scaler', StandardScaler())
])
# Create a preprocessor object
preprocessor = ColumnTransformer(
transformers=[
# ("cat", categorical_transformer, categorical_features),
("num", numeric_transformer, numeric_features)
])
# Create a pipeline that includes the preprocessor and the model
model = Pipeline(steps=[("preprocessor", preprocessor),
("model", RandomForestClassifier())])
# Split the data
X = df.drop("Crop", axis=1)
y = df["Crop"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Fit and score the model
model.fit(X_train, y_train)
model.score(X_test, y_test)
print('Baseline model:', model.score(X_test, y_test))
# Define the parameter grid
pipe_grid = {
'model__n_estimators': [10, 50, 100, 200],
'model__max_depth': [None, 10, 20, 30]
}
# Create a GridSearchCV object
gs_model = GridSearchCV(model, pipe_grid, cv=5, verbose=2, n_jobs=-1)
gs_model.fit(X_train, y_train)
print("Best parameters:", gs_model.best_params_)
print("Best score:", gs_model.best_score_)
# Fit the model with the entire training data using the best parameters
model.set_params(**gs_model.best_params_)
model.fit(X_train, y_train)
# Print the test score
print("Test score:", model.score(X_test, y_test))
Baseline model: 0.9931818181818182
Fitting 5 folds for each of 16 candidates, totalling 80 fits
Best parameters: {'model__max_depth': 30, 'model__n_estimators': 200}
Best score: 0.9960227272727273
Test score: 0.9931818181818182
In [ ]:
predictions = gs_model.best_estimator_.predict(X_test)
predictions